class: center, middle, inverse, title-slide # High-throughput Phenotyping Driven Quantitative Genetics <span class="citation">@CMA-FCT-NOVA</span> ## Structural equation model GWAS ### Gota Morota <br /><a href="http://morotalab.org/">http://morotalab.org/</a> <br /> ### 2021/10/22 --- # Acknowledgements This visit takes place within the scope of the Invited Researcher initiative of the Statistics and Risk Management group of CMA. It is funded by national funds through the FCT - Fundação para a Ciência e a Tecnologia, I.P., under the scope of the project UIDB/00297/2020 (Center for Mathematics and Applications). .pull-left[ <img src="NOVA.png" width=200 height=130> <img src="CMA.png" width=200 height=130> ] .pull-right[ <img src="NOVAID.png" width=200 height=100> <img src="FCT.png" width=200 height=120> <img src="RP.png" width=200 height=130> ] --- # Today's lunch .pull-left[ <img src="lunch1.jpg" width=350 height=350> Carvoada de Peixe ] .pull-right[ <img src="lunch2.jpg" width=350 height=350> View of Lisbon ] --- # Multivariate traits  - Multi-trait GWAS (MTM-GWAS) - Structural equation model GWAS (SEM-GWAS) --- # Multi-trait model $$ `\begin{align*} \mathbf{y} &= \mathbf{Xb} + \mathbf{Zu} + \mathbf{e} \\ \begin{bmatrix} \mathbf{y_1} \\ \mathbf{y_2} \\ \vdots \\ \mathbf{y_n} \\ \end{bmatrix} &= \begin{bmatrix} \mathbf{X_1} & 0 & \cdots & 0 \\ 0 & \mathbf{X_2} & \cdots & 0\\ \vdots & \vdots & & \vdots \\ 0 & 0 & \cdots & \mathbf{X_n} \\ \end{bmatrix} \begin{bmatrix} \mathbf{b_1} \\ \mathbf{b_2} \\ \vdots \\ \mathbf{b_n} \end{bmatrix} \\ &+ \begin{bmatrix} \mathbf{Z_1} & 0 & \cdots & 0 \\ 0 & \mathbf{Z_2} & \cdots & 0\\ \vdots & \vdots & & \vdots \\ 0 & 0 & \cdots & \mathbf{Z_n} \\ \end{bmatrix} \begin{bmatrix} \mathbf{u_1} \\ \mathbf{u_2} \\ \vdots \\ \mathbf{u_n} \end{bmatrix} + \begin{bmatrix} \mathbf{e_1} \\ \mathbf{e_2} \\ \vdots \\ \mathbf{e_n} \end{bmatrix} \\ E\begin{pmatrix} \mathbf{u} \\ \mathbf{e} \end{pmatrix} &= \begin{bmatrix} 0 \\ 0 \end{bmatrix} \\ Var\begin{pmatrix} \mathbf{u} \\ \mathbf{e} \end{pmatrix} &= \begin{bmatrix} \mathbf{\Sigma^2_{g}} \otimes \mathbf{G} & 0 \\ 0 & \mathbf{\Sigma^2_e} \otimes \mathbf{I} \end{bmatrix} \end{align*}` $$ --- # Multi-trait model (t = 2) $$ `\begin{align*} \mathbf{\Sigma^2_{g}} &= \begin{bmatrix} \mathbf{\Sigma^2_{g_{1}}} & \mathbf{\Sigma^2_{g_{12}}} \\ \mathbf{\Sigma^2_{g_{21}}} & \mathbf{\Sigma^2_{g_{2}}} \end{bmatrix} \\ \mathbf{\Sigma^2_{e}} &= \begin{bmatrix} \mathbf{\Sigma^2_{e_{1}}} & \mathbf{\Sigma^2_{e_{12}}} \\ \mathbf{\Sigma^2_{e_{21}}} & \mathbf{\Sigma^2_{e_{2}}} \end{bmatrix} \\ \mathbf{\Sigma^2_{g}} \otimes \mathbf{G} &= \begin{bmatrix} \mathbf{\Sigma^2_{g_{1}}} \mathbf{G} & \mathbf{\Sigma^2_{g_{12}}} \mathbf{G} \\ \mathbf{\Sigma^2_{g_{21}}} \mathbf{G} & \mathbf{\Sigma^2_{g_{2}}} \mathbf{G} \end{bmatrix} \\ \mathbf{\Sigma^2_{e}} \otimes \mathbf{I} &= \begin{bmatrix} \mathbf{\Sigma^2_{e_{1}}} \mathbf{I} & \mathbf{\Sigma^2_{e_{12}}} \mathbf{I} \\ \mathbf{\Sigma^2_{e_{21}}} \mathbf{I} & \mathbf{\Sigma^2_{e_{2}}} \mathbf{I} \end{bmatrix} \\ \end{align*}` $$ --- # Multi-trait model (t = 2) The MME model is $$ `\begin{align*} \begin{bmatrix} \mathbf{X'R_0^{-1}X} & \mathbf{X'R_0^{-1}Z} \\ \mathbf{Z'R_0^{-1}X} & \mathbf{Z'R_0^{-1}Z} + \mathbf{G_0}^{-1} \\ \end{bmatrix} \begin{bmatrix} \mathbf{b} \\ \mathbf{u} \end{bmatrix} &= \begin{bmatrix} \mathbf{X'R_0^{-1}y} \\ \mathbf{Z'R_0^{-1}y} \end{bmatrix} \\ \mathbf{G_0}^{-1} &= (\mathbf{\Sigma^2_{g}} \otimes \mathbf{G})^{-1} \\ &= \mathbf{\Sigma^2_{g}}^{-1} \otimes \mathbf{G}^{-1} \\ \mathbf{R_0}^{-1} &= (\mathbf{\Sigma^2_{e}} \otimes \mathbf{I})^{-1} \\ &= {\mathbf{\Sigma^2_{e}}}^{-1} \otimes \mathbf{I}^{-1} \end{align*}` $$ --- # Multi-trait GBLUP model `$$\mathbf{y_{1} = X_{1}b_{1} + Z_{1}u_{1} + e_{1}}$$` `$$\mathbf{y_{2} = X_{2}b_{2} + Z_{2}u_{2} + e_{2}}$$` Individuals within traits 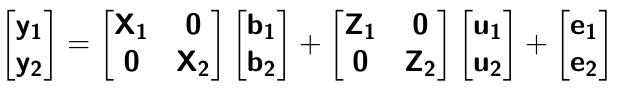 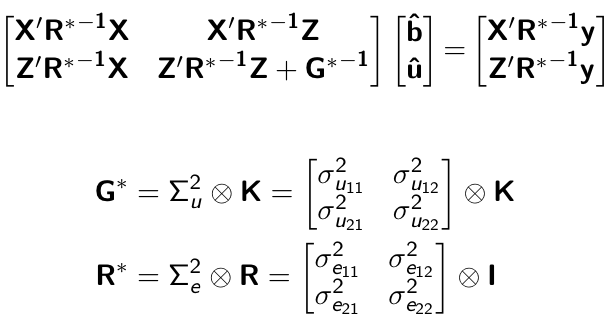 --- # Multi-trait RR-BLUP model `$$\mathbf{y_{1} = X_{1}b_{1} + W_{1}a_{1} + e_{1}}$$` `$$\mathbf{y_{2} = X_{2}b_{2} + W_{2}a_{2} + e_{2}}$$` Individuals within traits 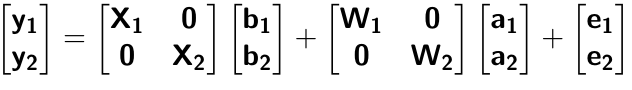 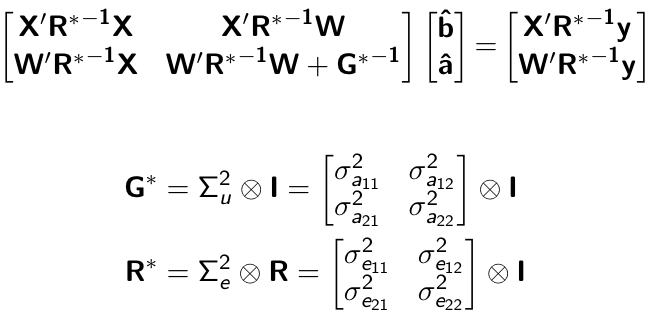 --- # Is multi-trait model useful? GBLUP model - estimate covariance structures `\(\rightarrow\)` genetic correlations - traits with scarce records - traits with low heritability RR-BLUP - increase statistical power to detect QTL regions --- # MTM-GWAS <img src="MTMGWAS.png" height="406px" width="650px"/> --- # Multiple-trait GWAS (MTM-GWAS) <img src="MTM.png" height="506px" width="750px"/> --- # Interrelationships among traits 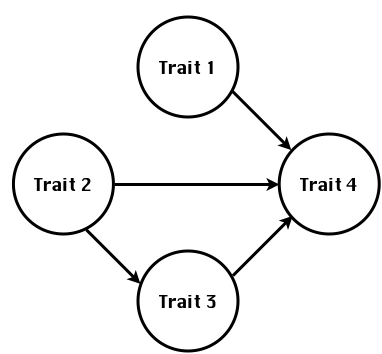 1. Biological prior - previous experiments - literature 2. Infer from the data --- # Phenotypic, genetic, and residual networks 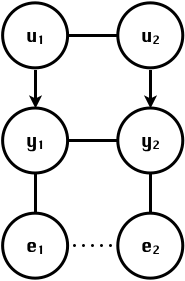 --- # Leveraging phenotypic networks to study multiple traits 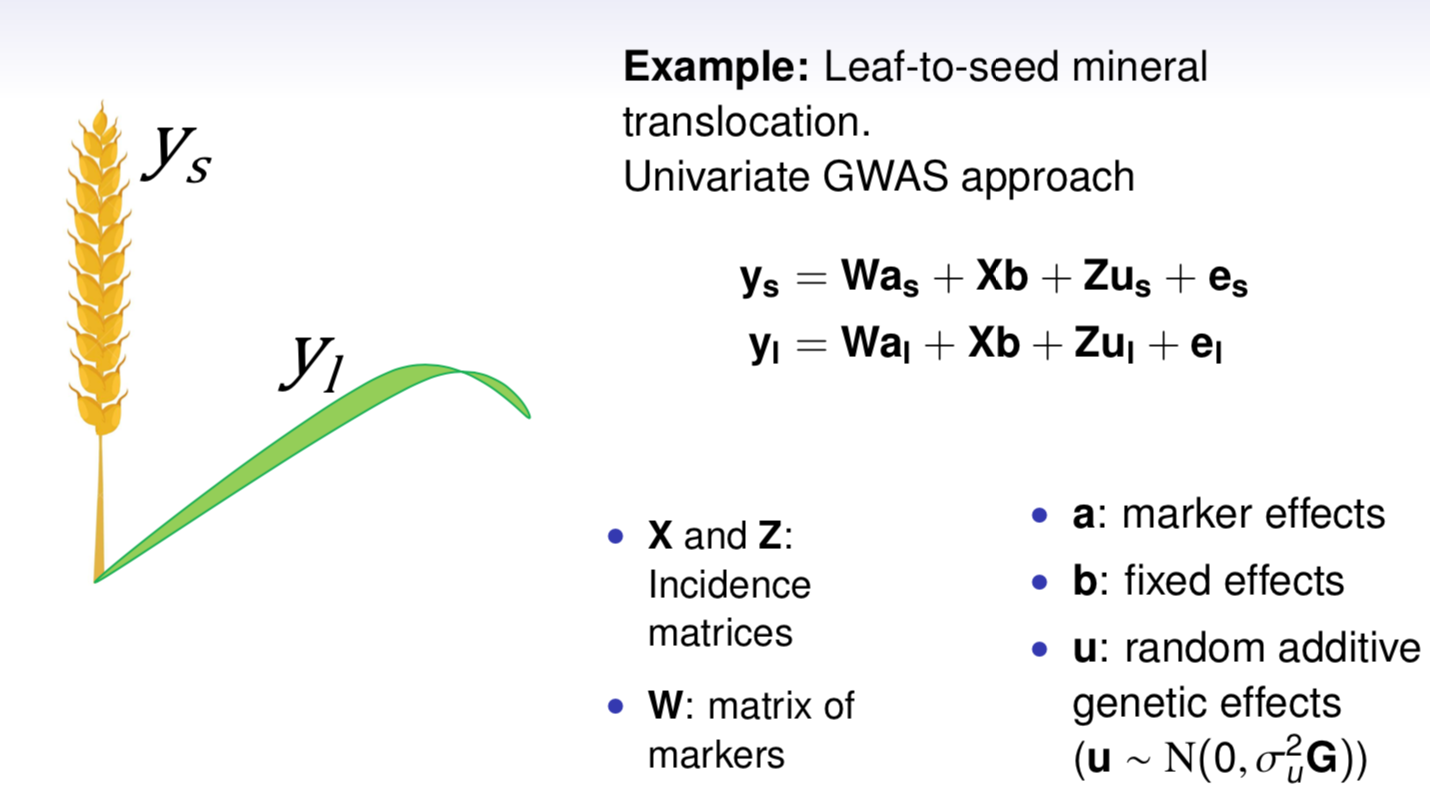 --- # Leveraging phenotypic networks to study multiple traits 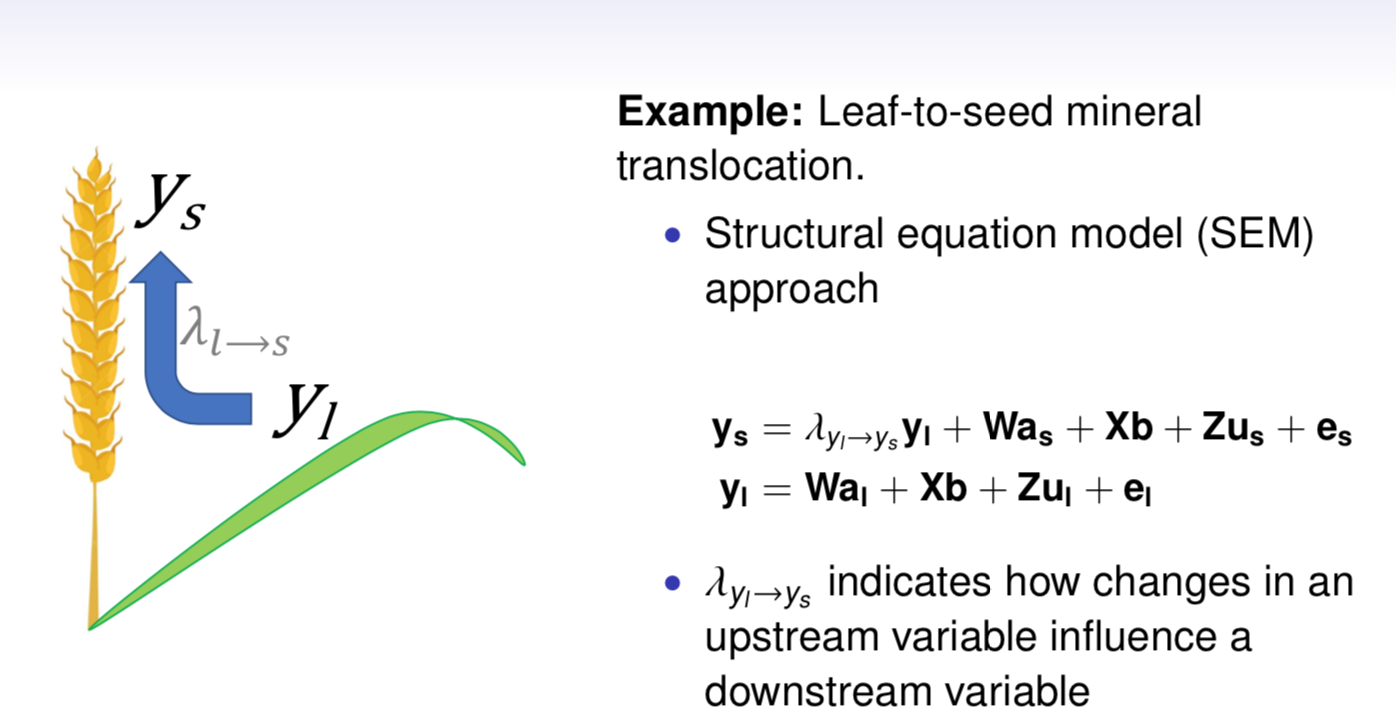 --- # Introducing SEM into the GWAS framework 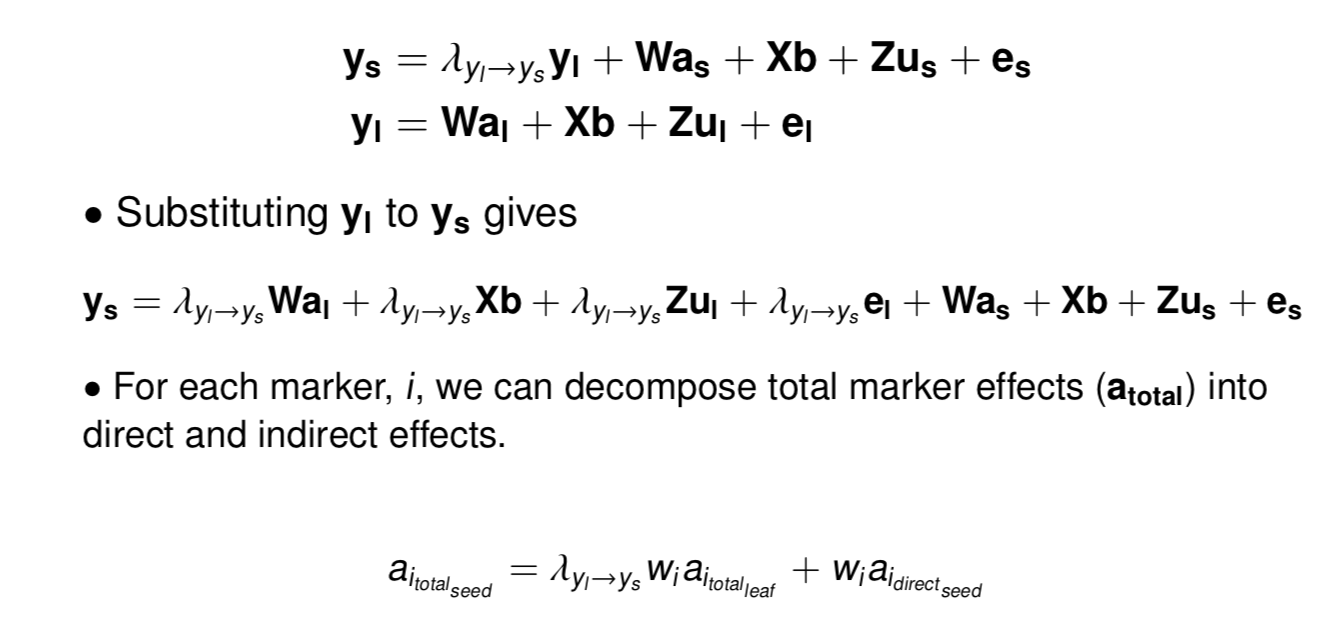 --- # Structural equation model GWAS (SEM-GWAS) Direct effect of SNP on Trait 4 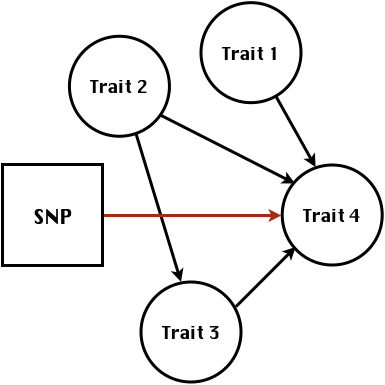 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 1 (1) 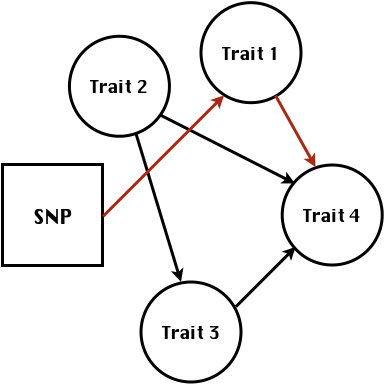 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 2 (2) 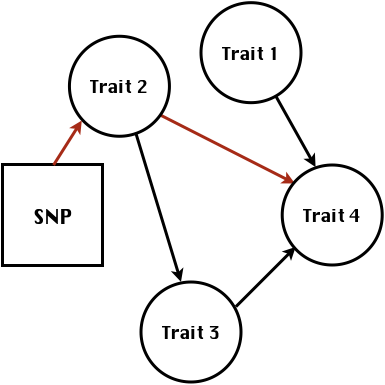 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 2 & 3 (3) 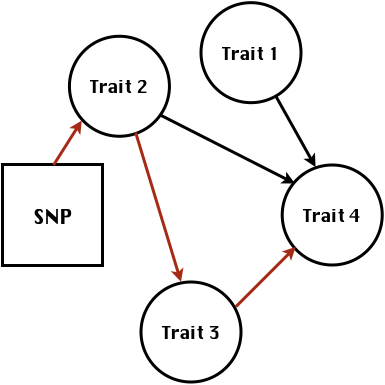 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 3 (4) 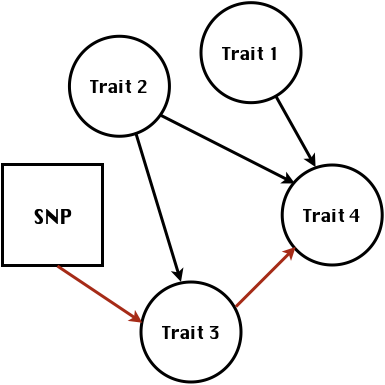 --- # SEM-GWAS vs. MTM-GWAS Effect of SNP on Trait 4 <img src="MTM4.png" height="306px" width="400px"/> `\begin{align*} \text{Effect of SNP (MTM-GWAS)} &\approx \text{Overall effect (SEM-GWAS)}\\ &= \text{Direct effect (SEM-GWAS)} \\ &+ \text{Indirect effect (SEM-GWAS)} \end{align*}` --- # How to fit SEM-GWAS? 1. Fit MTM and decompose phenotypes into genetics and residuals 2. Infer the network structure of traits 3. Fit SEM-GWAS given the network structure Example 1: Chicken data (n = 1,351), Momen et al. (2018) - breast muscle (BM) - body weight (BW) - hen-house egg production (HHP) --- # Inferred network structure (1) <img src="chickenSEM1.png" height="276px" width="600px"/> Structual equation model <img src="chickenSEM5.png" height="166px" width="500px"/> --- # Inferred network structure (2) <img src="chickenSEM1.png" height="276px" width="600px"/> Structual equation model <img src="chickenSEM3.png" height="56px" width="250px"/> <img src="chickenSEM4.png" height="106px" width="500px"/> --- # Inferred network structure (3) <img src="chickenSEM1.png" height="276px" width="600px"/> Decomposition of SNP effects <img src="chickenSEM2.png" height="176px" width="500px"/> --- # Manhattan plots of BW (overall, direct, and indirect SNP effects) <img src="chickenSEM6.png" height="426px" width="560px"/> --- # More details about Chicken SEM-GWAS <img src="frontiers.png" height="400px" width="630px"/> --- # Leveraging phenotypic networks to study multiple traits  --- # Leveraging phenotypic networks to study multiple traits  .pull-left[ - Shoot biomass (projected shoot area, PSA) - Root biomass (RB) ] .pull-right[ - Water use (WU) - Water use efficiency (WUE) ] How are these traits related? --- # Rice data - Drought data - Rice diversity panel - Projected shoot area - Root biomass - Water use efficiency - Water use Inferred trait network structure <img src="riceSEM1.png" height="326px" width="460px"/> --- # Bayesian networks .pull-left[  ] .pull-right[ - BN based on 500 bootstrap samples - % above edges indicate percentage of bootstrap networks with the given edge - Numbers in parenthesis indicate the proportion of bootstrap networks with the given edge direction ] --- # Bayesian networks .pull-left[  ] .pull-right[ - Phenotypic values for WUE are dependant on phenotypes for RB and PSA - Phenotypic values for WU are dependant on phenotypes for PSA ] --- # How to fit SEM-GWAS? `\begin{align*} \mathbf{y} =\boldsymbol{\Lambda} \mathbf{y} + \mathbf{ws} + \mathbf{Zg} + \boldsymbol{\epsilon} \end{align*}` <img src="riceSEM2.png" height="426px" width="600px"/> --- # Inferred trait network structure <img src="riceSEM3.png" height="426px" width="700px"/> --- # Structural equation model <img src="riceSEM4.png" height="426px" width="700px"/> --- # Bayesian information criterion <img src="riceSEM10.png" height="306px" width="620px"/> --- # Structural coefficients <img src="riceSEM11.png" height="306px" width="610px"/> --- # Projected shoot area `\begin{align*} \text{Direct}_{s_j \rightarrow y_{1_{\text{PSA}}}} &= s_{j(y_{1_{\text{PSA}}})} \\ \text{Total}_{s_j \rightarrow y_{1_{\text{PSA}}}} &= \text{Direct}_{s_j \rightarrow y_{1_{\text{PSA}}}}\\ &= s_{j(y_{1_{\text{PSA}}})} \end{align*}` <img src="riceSEM5.png" height="406px" width="700px"/> --- # Root biomass `\begin{align*} \text{Direct}_{s_j \rightarrow y_{2_{\text{RB}}}} &=s_{j(y_{2_{\text{RB}}})} \\ \text{Total}_{s_j \rightarrow y_{2_{\text{RB}}}} &= \text{Direct}_{s_j \rightarrow y_{2_{\text{RB}}}}\\ &= s_{j(y_{2_{\text{RB}}})} \end{align*}` <img src="riceSEM5.png" height="406px" width="700px"/> --- # Water use `\begin{align*} \text{Direct}_{s_j \rightarrow y_{3_{\text{WU}}}} &=s_{j(y_{3_{\text{WU}}})} \\ \text{Indirect}_{s_j \rightarrow y_{3_{\text{WU}}}} &= \lambda_{13}s_{j(y_{1_{\text{PSA}}})} \\ \text{Total}_{s_j \rightarrow y_{3_{\text{WU}}}} &= \text{Direct}_{s_j \rightarrow y_{3_{\text{WU}}}} + \text{Indirect}_{s_j \rightarrow y_{3_{\text{WU}}}} \\ &= s_{j(y_{3_{\text{WU}}})} + \lambda_{13}s_{j(y_{1_{\text{PSA}}})} \end{align*}` <img src="riceSEM7.png" height="336px" width="700px"/> --- # Water use efficiency `\begin{align*} \text{Direct}_{s_j \rightarrow y_{4_{\text{WUE}}}} &=s_{j(y_{4_{\text{WUE}}})} \\ \text{Indirect(1)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{14}s_{j(y_{1_{\text{PSA}}})} \\ \text{Indirect(2)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{24}s_{j(y_{2_{\text{RB}}})} \\ \text{Total}_{S_j \rightarrow y_{4_{\text{WUE}}}} &= \text{Direct}_{s_j \rightarrow y_{4_{\text{WUE}}}} + \text{Indirect(1)}_{s_j \rightarrow y_{4_{\text{WUE}}}} + \text{Indirect(2)}_{s_j \rightarrow y_{4_{\text{WUE}}}} \\ &= s_{j(y_{4_{\text{WUE}}})} + \lambda_{14}s_{j(y_{1_{\text{PSA}}})} + \lambda_{24}s_{j(y_{2_{\text{RB}}})} \end{align*}` --- # Water use efficiency <img src="riceSEM8.png" height="536px" width="700px"/> --- # Assumption about marker effects - BLUP assumes normally distributed QTL / marker effects - Does not match the prior information regarding the distributions of QTL / marker effects for some traits - What if we want to assume QTL / markers have non-Gaussian distribution? --- # BayesRR (ridge regression) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a} \sim N(0, \sigma_{a}^{2} ) \\ \sigma^2_{a} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesL (Lasso) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \\ \sigma^2_{a_j} \sim Exponential(\lambda) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` --- # BayesA `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesB `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \\ \begin{cases} \sigma^2_{a_j} = 0 \quad \text{with probability} \quad \pi\\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesB* (modified version) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j = 0 \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesB** (modified version) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} \approx 0) \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesCpi `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j = 0 \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a} \sim N(0, \sigma_{a}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \pi \sim Uniform(0,1) \\ \sigma^2_{a} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesC `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j = 0 \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a} \sim N(0, \sigma_{a}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{a} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # Prior densities ([de los Campos et al., 2012](https://doi.org/10.1534/genetics.112.143313)) <img src="deloscampos2012.png" width=700 height=280> .pull-left[ - Bayes RR (ridge regression) - BayesL (LASSO) - BayesA - BayesB ] .pull-right[ - BayesB* - BayesB** - BayesC - BayesCpi ] --- # Bayesian marker effect SEM 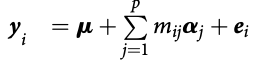 - `\(\mathbf{\alpha_j} = \mathbf{D_j B_j}\)` - `\(\mathbf{D_j}\)`: diagonal matrix including the indicator variable indicating whether the marker effect of locus j for trait k is zero or not - `\(\mathbf{B_j}\)`: a priori assumed to be independently and identically distributed multivariate normal vectors  - `\(\boldsymbol{\lambda}\)`: non-zero elements in `\(\boldsymbol{\Lambda}\)` --- # Distribution for `\(\lambda\)` <small>Prior </small> 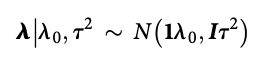 <small>Full conditional </small> <img src="BSEM3.png" height="290px" width="460px"/> - `\(\lambda_0 = 0\)`, `\(\tau = 1\)`, and `\(\mathbf{R}\)` is the residual covariance matrix --- # Simulated data 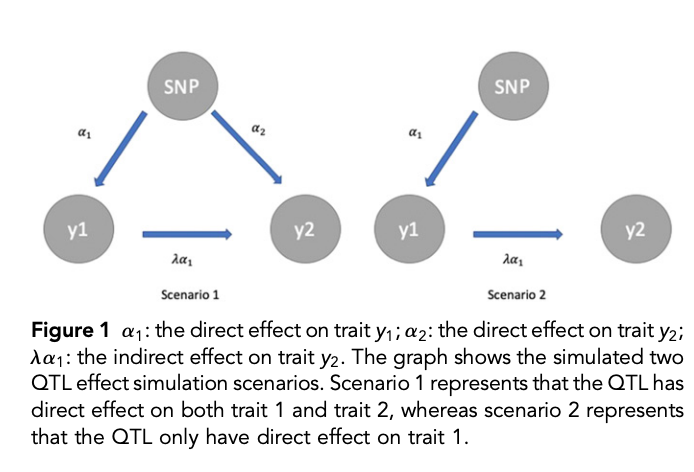 --- # Simulated data - Results 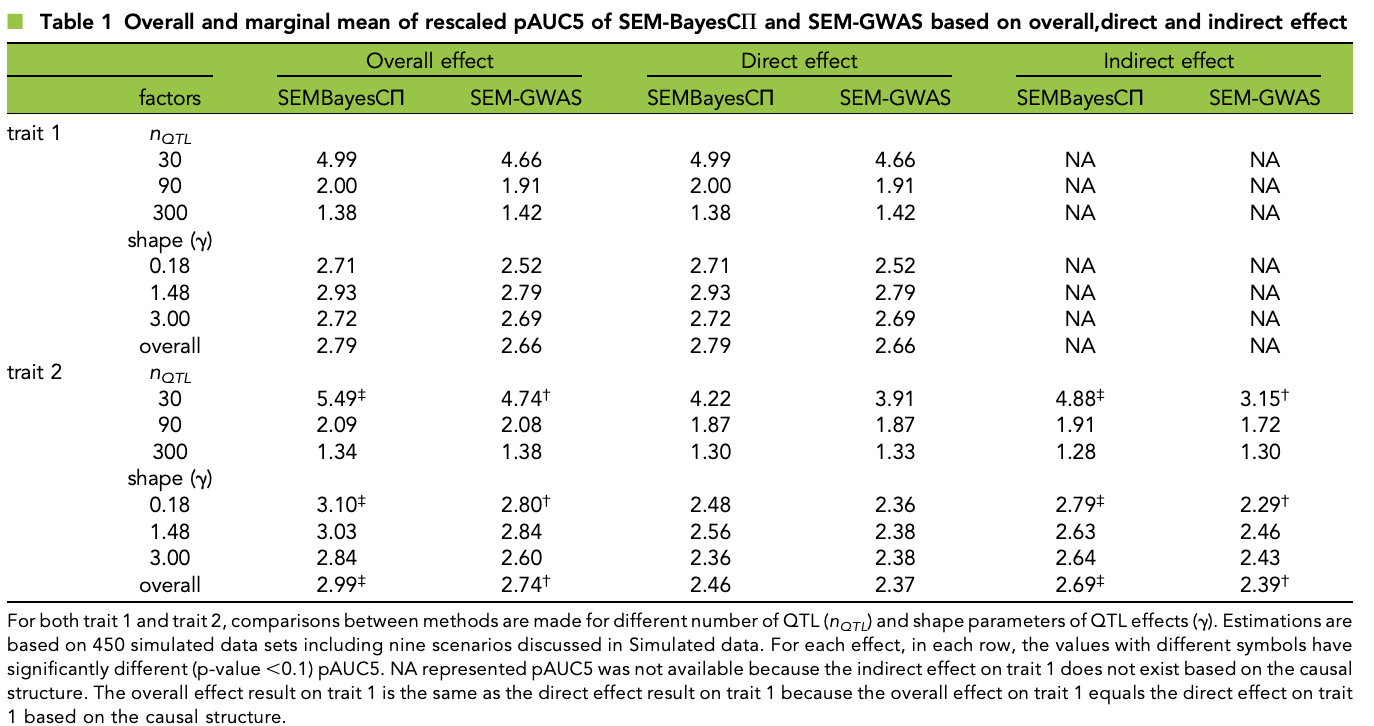 --- # Real rice data 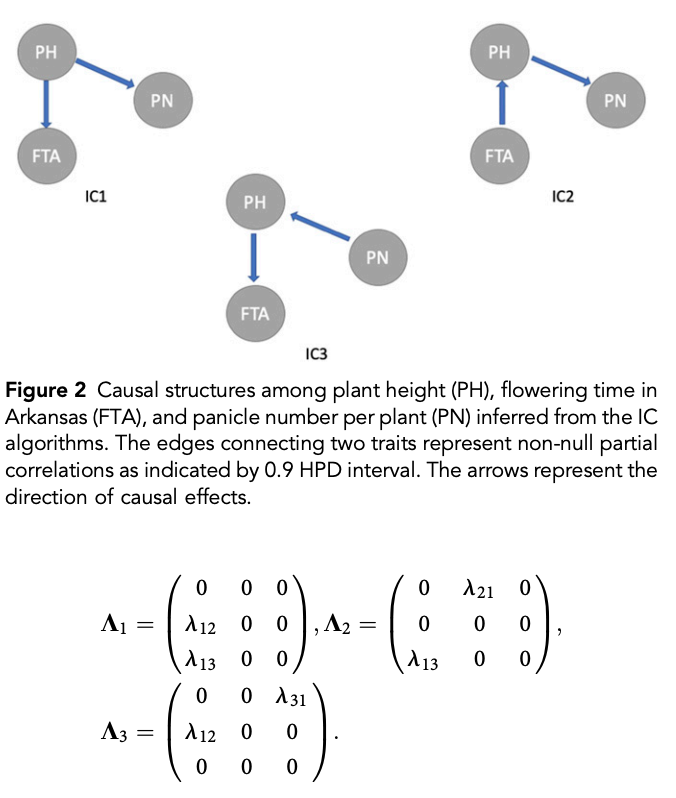 --- # Real rice data - Resutls  --- # Reference  --- # Conclusions for SEM-GWAS - Relatively simple approach to understand how QTL affect multiple traits - Provides novel insights compared to conventional multi-trait GWAS approaches