class: center, middle, inverse, title-slide # Structural equation model GWAS - Advanced GWAS approaches <span class="citation">@IRRI-NARES</span> ## IRRI Virtual Training Program: Breeding Innovation for Crop Improvement to Enhance Genetic Gains ### Gota Morota <br /><a href="http://morotalab.org/">http://morotalab.org/</a> <br /> ### 2021/11/12 --- # About me  --- # Multivariate traits  - Multi-trait GWAS (MTM-GWAS) - Structural equation model GWAS (SEM-GWAS) --- # Multi-trait model $$ `\begin{align*} \mathbf{y} &= \mathbf{Xb} + \mathbf{Zu} + \mathbf{e} \\ \begin{bmatrix} \mathbf{y_1} \\ \mathbf{y_2} \\ \vdots \\ \mathbf{y_n} \\ \end{bmatrix} &= \begin{bmatrix} \mathbf{X_1} & 0 & \cdots & 0 \\ 0 & \mathbf{X_2} & \cdots & 0\\ \vdots & \vdots & & \vdots \\ 0 & 0 & \cdots & \mathbf{X_n} \\ \end{bmatrix} \begin{bmatrix} \mathbf{b_1} \\ \mathbf{b_2} \\ \vdots \\ \mathbf{b_n} \end{bmatrix} \\ &+ \begin{bmatrix} \mathbf{Z_1} & 0 & \cdots & 0 \\ 0 & \mathbf{Z_2} & \cdots & 0\\ \vdots & \vdots & & \vdots \\ 0 & 0 & \cdots & \mathbf{Z_n} \\ \end{bmatrix} \begin{bmatrix} \mathbf{u_1} \\ \mathbf{u_2} \\ \vdots \\ \mathbf{u_n} \end{bmatrix} + \begin{bmatrix} \mathbf{e_1} \\ \mathbf{e_2} \\ \vdots \\ \mathbf{e_n} \end{bmatrix} \\ E\begin{pmatrix} \mathbf{u} \\ \mathbf{e} \end{pmatrix} &= \begin{bmatrix} 0 \\ 0 \end{bmatrix} \\ Var\begin{pmatrix} \mathbf{u} \\ \mathbf{e} \end{pmatrix} &= \begin{bmatrix} \mathbf{\Sigma^2_{g}} \otimes \mathbf{G} & 0 \\ 0 & \mathbf{\Sigma^2_e} \otimes \mathbf{I} \end{bmatrix} \end{align*}` $$ --- # Multi-trait model (t = 2) $$ `\begin{align*} \mathbf{\Sigma^2_{g}} &= \begin{bmatrix} \mathbf{\Sigma^2_{g_{1}}} & \mathbf{\Sigma^2_{g_{12}}} \\ \mathbf{\Sigma^2_{g_{21}}} & \mathbf{\Sigma^2_{g_{2}}} \end{bmatrix} \\ \mathbf{\Sigma^2_{e}} &= \begin{bmatrix} \mathbf{\Sigma^2_{e_{1}}} & \mathbf{\Sigma^2_{e_{12}}} \\ \mathbf{\Sigma^2_{e_{21}}} & \mathbf{\Sigma^2_{e_{2}}} \end{bmatrix} \\ \mathbf{\Sigma^2_{g}} \otimes \mathbf{G} &= \begin{bmatrix} \mathbf{\Sigma^2_{g_{1}}} \mathbf{G} & \mathbf{\Sigma^2_{g_{12}}} \mathbf{G} \\ \mathbf{\Sigma^2_{g_{21}}} \mathbf{G} & \mathbf{\Sigma^2_{g_{2}}} \mathbf{G} \end{bmatrix} \\ \mathbf{\Sigma^2_{e}} \otimes \mathbf{I} &= \begin{bmatrix} \mathbf{\Sigma^2_{e_{1}}} \mathbf{I} & \mathbf{\Sigma^2_{e_{12}}} \mathbf{I} \\ \mathbf{\Sigma^2_{e_{21}}} \mathbf{I} & \mathbf{\Sigma^2_{e_{2}}} \mathbf{I} \end{bmatrix} \\ \end{align*}` $$ --- # Multi-trait GBLUP model `$$\mathbf{y_{1} = X_{1}b_{1} + Z_{1}u_{1} + e_{1}}$$` `$$\mathbf{y_{2} = X_{2}b_{2} + Z_{2}u_{2} + e_{2}}$$` Individuals within traits 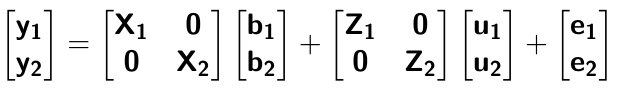 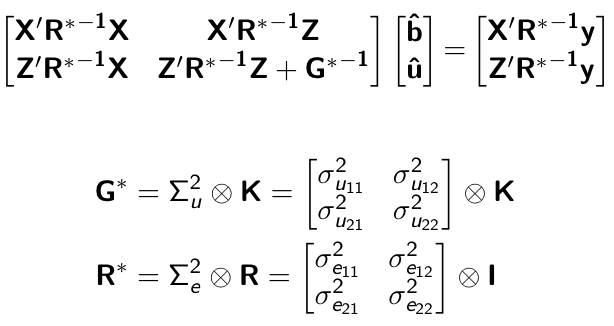 --- # Is multi-trait model useful? - increase statistical power to detect QTL regions - estimate covariance structures `\(\rightarrow\)` genetic correlations - traits with scarce records - traits with low heritability --- # MTM-GWAS (Single-marker linear mixed model) <img src="MTMGWAS.png" height="406px" width="650px"/> --- # Multiple-trait GWAS (MTM-GWAS) <img src="MTM.png" height="506px" width="750px"/> --- # Interrelationships among traits  1. Biological prior - previous experiments - literature 2. Infer from the data --- # Bayesian Network Probabilistic directed acyclic graphical model - interrelationship (Edges) among latent variables (Nodes) - genetic selection for breeding requires causal assumptions <div align="center"> <img src="BCFA2.png" height="256px" width="400px"/> </div> --- # Constraint-based learning <img src="Fig3.jpg" height="530px" width="700px"/> --- # Score-based learning <img src="Score-based-diagram.png" height="500px" width="700px"/> --- # Examples of algorithms Score-based algorithm - Hill climbing - Tabu search Hybrid algorithm - Max-Min Hill Climbing algorithm - General 2-Phase Restricted Maximization algorithm --- # References <img src="G32018.png" height="230px" width="500px"/> [10.1534/g3.119.400154](https://doi.org/10.1534/g3.119.400154) <img src="momen.png" height="230px" width="550px"/> [10.1002/pld3.304](https://doi.org/10.1002/pld3.304) --- # Phenotypic, genetic, and residual networks  --- # Leveraging phenotypic networks to study multiple traits  --- # Leveraging phenotypic networks to study multiple traits  --- # Introducing SEM into the GWAS framework 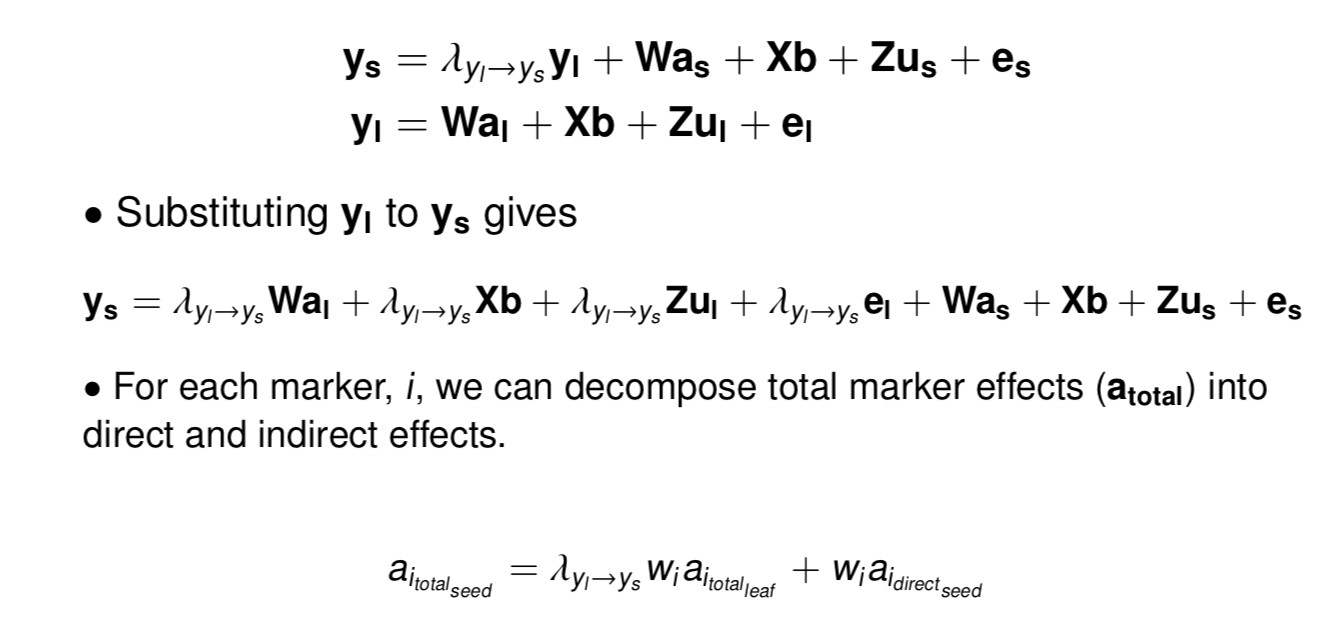 --- # Structural equation model GWAS (SEM-GWAS) Direct effect of SNP on Trait 4 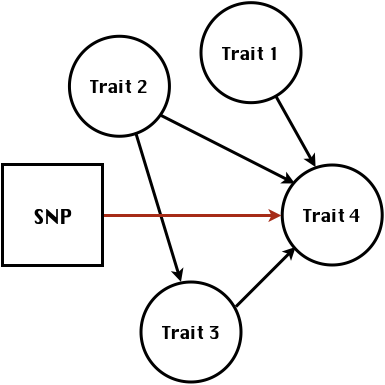 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 1 (1) 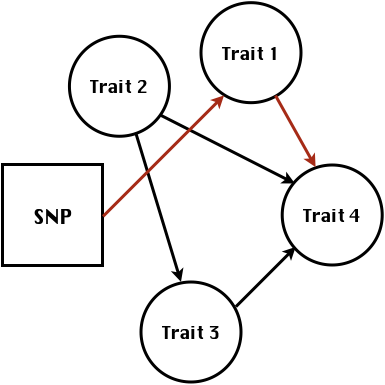 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 2 (2) 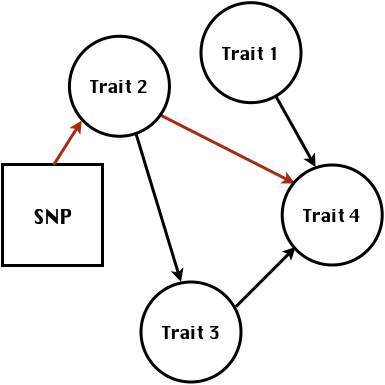 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 2 & 3 (3)  --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 3 (4)  --- # SEM-GWAS vs. MTM-GWAS Effect of SNP on Trait 4 <img src="MTM4.png" height="306px" width="400px"/> `\begin{align*} \text{Effect of SNP (MTM-GWAS)} &\approx \text{Overall effect (SEM-GWAS)}\\ &= \text{Direct effect (SEM-GWAS)} \\ &+ \text{Indirect effect (SEM-GWAS)} \end{align*}` --- # How to fit SEM-GWAS? 1. Fit MTM and decompose phenotypes into genetics and residuals 2. Infer the network structure of traits 3. Fit SEM-GWAS given the network structure --- # Leveraging phenotypic networks to study multiple traits  --- # Leveraging phenotypic networks to study multiple traits  .pull-left[ - Shoot biomass (projected shoot area, PSA) - Root biomass (RB) ] .pull-right[ - Water use (WU) - Water use efficiency (WUE) ] How are these traits related? --- # Projected shoot area <img src="PSA5.png" height="530px" width="600px"/> --- # Rice data - Drought data - Rice diversity panel - Projected shoot area - Root biomass - Water use efficiency - Water use Inferred trait network structure <img src="riceSEM1.png" height="326px" width="460px"/> --- # Bayesian networks .pull-left[  ] .pull-right[ - BN based on 500 bootstrap samples - % above edges indicate percentage of bootstrap networks with the given edge - Numbers in parenthesis indicate the proportion of bootstrap networks with the given edge direction ] --- # Bayesian networks .pull-left[  ] .pull-right[ - Phenotypic values for WUE are dependant on phenotypes for RB and PSA - Phenotypic values for WU are dependant on phenotypes for PSA ] --- # How to fit SEM-GWAS? `\begin{align*} \mathbf{y} =\boldsymbol{\Lambda} \mathbf{y} + \mathbf{ws} + \mathbf{Zg} + \boldsymbol{\epsilon} \end{align*}` <img src="riceSEM2.png" height="426px" width="600px"/> --- # Inferred trait network structure <img src="riceSEM3.png" height="426px" width="700px"/> --- # Structural equation model <img src="riceSEM4.png" height="426px" width="700px"/> --- # Bayesian information criterion <img src="riceSEM10.png" height="306px" width="620px"/> --- # Structural coefficients <img src="riceSEM11.png" height="306px" width="610px"/> --- # Projected shoot area `\begin{align*} \text{Direct}_{s_j \rightarrow y_{1_{\text{PSA}}}} &= s_{j(y_{1_{\text{PSA}}})} \\ \text{Total}_{s_j \rightarrow y_{1_{\text{PSA}}}} &= \text{Direct}_{s_j \rightarrow y_{1_{\text{PSA}}}}\\ &= s_{j(y_{1_{\text{PSA}}})} \end{align*}` <img src="riceSEM5.png" height="406px" width="700px"/> --- # Root biomass `\begin{align*} \text{Direct}_{s_j \rightarrow y_{2_{\text{RB}}}} &=s_{j(y_{2_{\text{RB}}})} \\ \text{Total}_{s_j \rightarrow y_{2_{\text{RB}}}} &= \text{Direct}_{s_j \rightarrow y_{2_{\text{RB}}}}\\ &= s_{j(y_{2_{\text{RB}}})} \end{align*}` <img src="riceSEM5.png" height="406px" width="700px"/> --- # Water use `\begin{align*} \text{Direct}_{s_j \rightarrow y_{3_{\text{WU}}}} &=s_{j(y_{3_{\text{WU}}})} \\ \text{Indirect}_{s_j \rightarrow y_{3_{\text{WU}}}} &= \lambda_{13}s_{j(y_{1_{\text{PSA}}})} \\ \text{Total}_{s_j \rightarrow y_{3_{\text{WU}}}} &= \text{Direct}_{s_j \rightarrow y_{3_{\text{WU}}}} + \text{Indirect}_{s_j \rightarrow y_{3_{\text{WU}}}} \\ &= s_{j(y_{3_{\text{WU}}})} + \lambda_{13}s_{j(y_{1_{\text{PSA}}})} \end{align*}` <img src="riceSEM7.png" height="336px" width="700px"/> --- # Water use efficiency `\begin{align*} \text{Direct}_{s_j \rightarrow y_{4_{\text{WUE}}}} &=s_{j(y_{4_{\text{WUE}}})} \\ \text{Indirect(1)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{14}s_{j(y_{1_{\text{PSA}}})} \\ \text{Indirect(2)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{24}s_{j(y_{2_{\text{RB}}})} \\ \text{Total}_{S_j \rightarrow y_{4_{\text{WUE}}}} &= \text{Direct}_{s_j \rightarrow y_{4_{\text{WUE}}}} + \text{Indirect(1)}_{s_j \rightarrow y_{4_{\text{WUE}}}} + \text{Indirect(2)}_{s_j \rightarrow y_{4_{\text{WUE}}}} \\ &= s_{j(y_{4_{\text{WUE}}})} + \lambda_{14}s_{j(y_{1_{\text{PSA}}})} + \lambda_{24}s_{j(y_{2_{\text{RB}}})} \end{align*}` --- # Water use efficiency <img src="riceSEM8.png" height="536px" width="700px"/> --- # Assumption about marker effects - BLUP assumes normally distributed QTL / marker effects - Does not match the prior information regarding the distributions of QTL / marker effects for some traits - What if we want to assume QTL / markers have non-Gaussian distribution? --- # BayesRR (ridge regression) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a} \sim N(0, \sigma_{a}^{2} ) \\ \sigma^2_{a} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesL (Lasso) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \\ \sigma^2_{a_j} \sim Exponential(\lambda) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` --- # BayesA `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesB `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \\ \begin{cases} \sigma^2_{a_j} = 0 \quad \text{with probability} \quad \pi\\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesB* (modified version) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j = 0 \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesB** (modified version) `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} \approx 0) \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a_j} \sim N(0, \sigma_{a_j}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{a_j} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesCpi `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j = 0 \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a} \sim N(0, \sigma_{a}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \pi \sim Uniform(0,1) \\ \sigma^2_{a} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # BayesC `\begin{align*} \mathbf{y} = \sum^m_{j=1} \mathbf{W}_{j} a_j + \epsilon \rightarrow \mathbf{y}| a_j, \sigma^2_e \sim N(\mathbf{W}_{j} a_j, \sigma^2_e) \end{align*}` `\(p(\mathbf{a}| \mathbf{y}, \sigma^2_e, \sigma^2_{a}) \propto p(\mathbf{y}|\mathbf{a}, \sigma^2_e) \cdot p(\mathbf{a}| \sigma^2_{a})\)` `\begin{align*} \text{Prior distributions} \begin{cases} \begin{cases} a_j = 0 \quad \text{with probability} \quad \pi \\ a_j|\sigma^2_{a} \sim N(0, \sigma_{a}^{2} ) \quad \text{with probability} \quad (1 - \pi) \end{cases} \\ \sigma^2_{a} \sim \chi^{-2}(\nu, S) \\ \sigma^2_{e} \sim \chi^{-2}(\nu, S) \end{cases} \end{align*}` - `\(\pi\)`: probability that markers have no effect - `\(\chi^{-2}(\nu, S) \rightarrow\)` scaled inverted chi-square distribution with `\(\nu\)` degrees of freedom and scale parameter `\(S\)` --- # Prior densities ([de los Campos et al., 2012](https://doi.org/10.1534/genetics.112.143313)) <img src="deloscampos2012.png" width=700 height=280> .pull-left[ - Bayes RR (ridge regression) - BayesL (LASSO) - BayesA - BayesB ] .pull-right[ - BayesB* - BayesB** - BayesC - BayesCpi ] --- # Bayesian marker effect SEM  - `\(\mathbf{\alpha_j} = \mathbf{D_j B_j}\)` - `\(\mathbf{D_j}\)`: diagonal matrix including the indicator variable indicating whether the marker effect of locus j for trait k is zero or not - `\(\mathbf{B_j}\)`: a priori assumed to be independently and identically distributed multivariate normal vectors  - `\(\boldsymbol{\lambda}\)`: non-zero elements in `\(\boldsymbol{\Lambda}\)` --- # Distribution for `\(\lambda\)` <small>Prior </small>  <small>Full conditional </small> <img src="BSEM3.png" height="290px" width="460px"/> - `\(\lambda_0 = 0\)`, `\(\tau = 1\)`, and `\(\mathbf{R}\)` is the residual covariance matrix --- # Real rice data 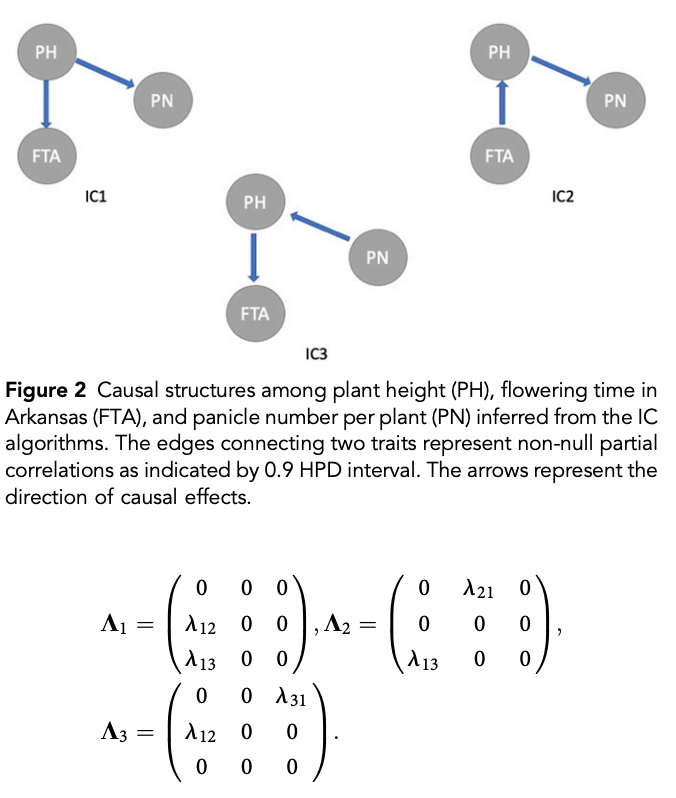 --- # Real rice data - Resutls 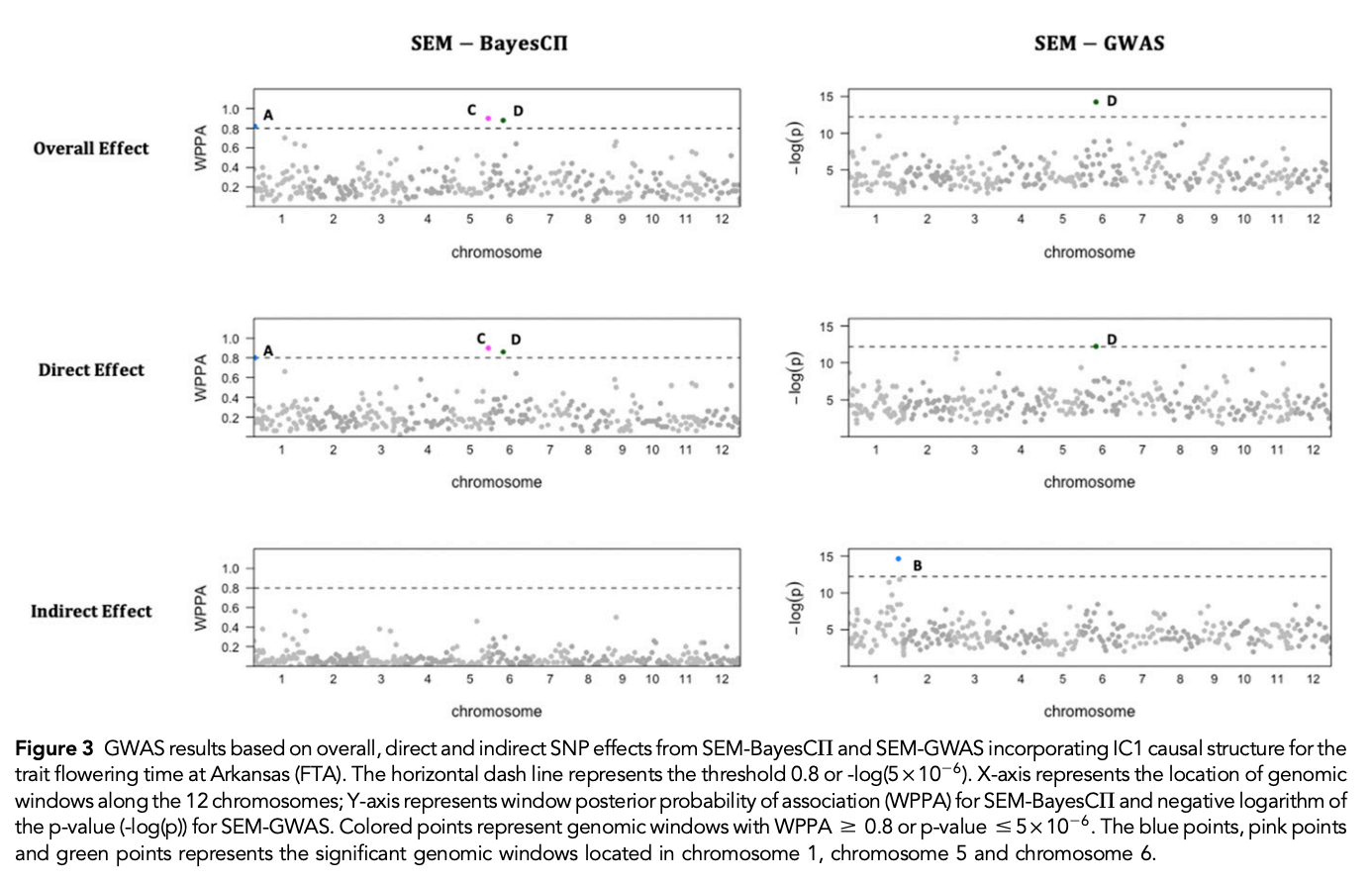 --- # Reference  [10.1534/g3.120.401618](https://doi.org/10.1534/g3.120.401618) --- # Implementation of Bayesian marker effect SEM-GWAS 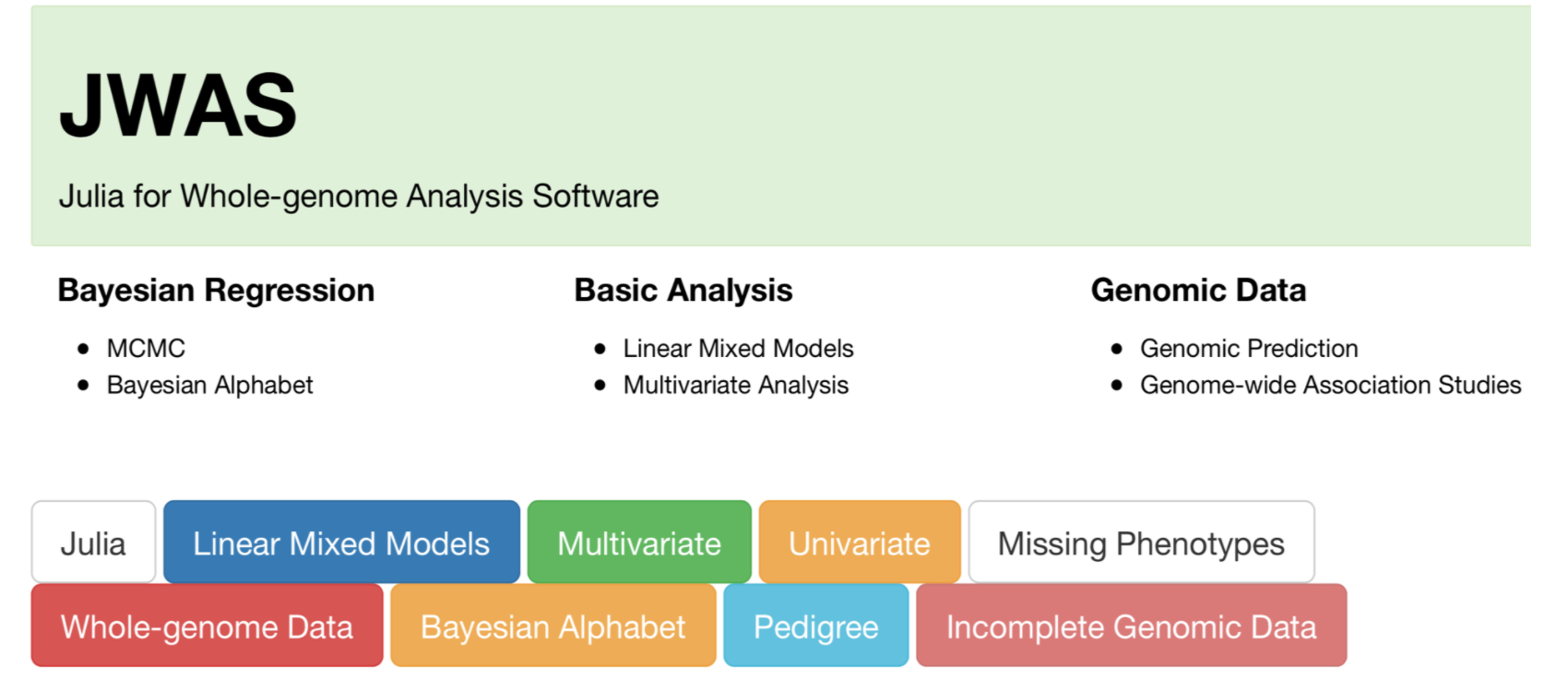 [https://reworkhow.github.io/JWAS.jl/latest/](https://reworkhow.github.io/JWAS.jl/latest/) You can fit BayesA SEM-GWAS, BayesB SEM-GWAS, BayesCpi SEM-GWAS, etc.! --- # Conclusions for SEM-GWAS - Relatively simple approach to understand how QTL affect multiple traits - Provides novel insights compared to conventional multi-trait GWAS approaches