class: center, middle, inverse, title-slide # Statistical methods for the quantitative genetic analysis of high-throughput phenotyping data ## Structural equation model GWAS <br /> The 64th RBras and 18th SEAGRO Meeting ### Gota Morota <br /><a href="http://morotalab.org/" class="uri">http://morotalab.org/</a> <br /> ### 2019/8/2 --- # Multivariate traits  - Multi-trait GWAS (MTM-GWAS) - Structural equation model GWAS (SEM-GWAS) --- # MTM-GWAS <img src="MTMGWAS.png" height="406px" width="650px"/> --- # Multiple-trait GWAS (MTM-GWAS) <img src="MTM.png" height="506px" width="750px"/> --- # Interrelationships among traits 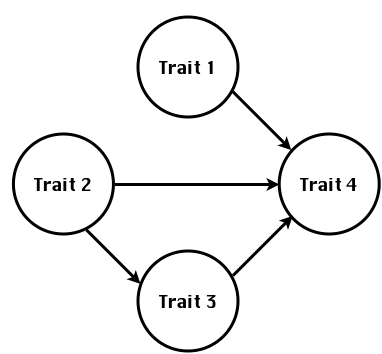 1. Biological prior - previous experiments - literature 2. Infer from the data --- # Phenotypic, genetic, and residual networks 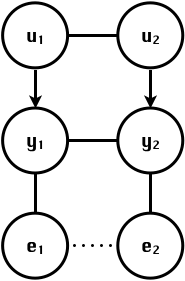 --- # Structural equation model GWAS (SEM-GWAS) Direct effect of SNP on Trait 4 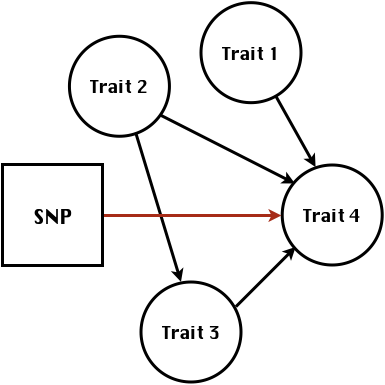 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 1 (1) 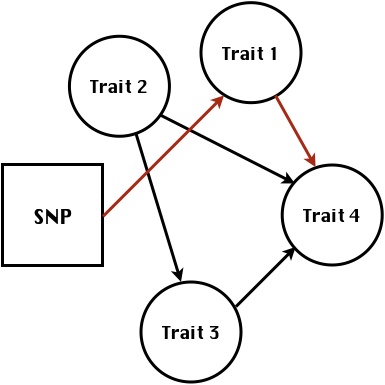 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 2 (2) 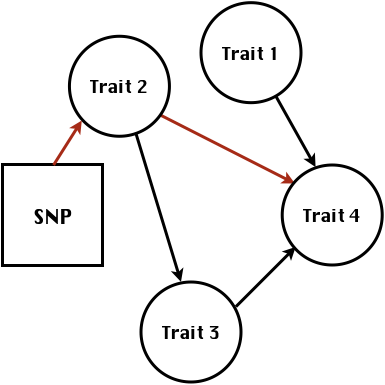 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 2 & 3 (3) 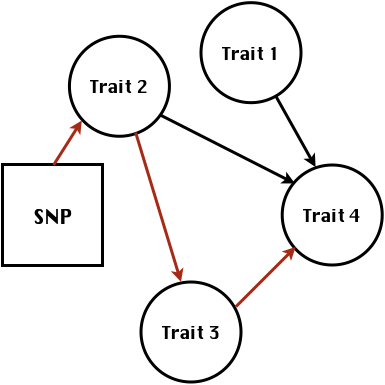 --- # SEM-GWAS Indirect effect of SNP on Trait 4 through Trait 3 (4) 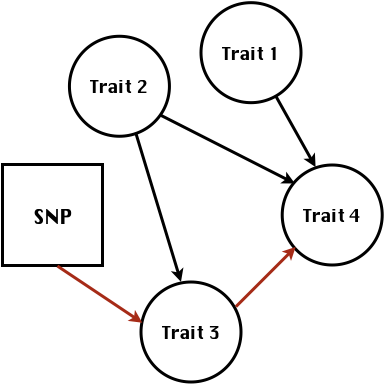 --- # SEM-GWAS vs. MTM-GWAS Effect of SNP on Trait 4 <img src="MTM4.png" height="306px" width="400px"/> `\begin{align*} \text{Effect of SNP (MTM-GWAS)} &\approx \text{Overall effect (SEM-GWAS)}\\ &= \text{Direct effect (SEM-GWAS)} \\ &+ \text{Indirect effect (SEM-GWAS)} \end{align*}` --- # How to fit SEM-GWAS? 1. Fit MTM and decompose phenotypes into genetics and residuals 2. Infer the network structure of traits 3. Fit SEM-GWAS given the network structure Example 1: Chicken data (n = 1,351), Momen et al. (2018) - breast muscle (BM) - body weight (BW) - hen-house egg production (HHP) --- # Inferred network structure (1) <img src="chickenSEM1.png" height="276px" width="600px"/> Structual equation model <img src="chickenSEM5.png" height="166px" width="500px"/> --- # Inferred network structure (2) <img src="chickenSEM1.png" height="276px" width="600px"/> Structual equation model <img src="chickenSEM3.png" height="56px" width="250px"/> <img src="chickenSEM4.png" height="106px" width="500px"/> --- # Inferred network structure (3) <img src="chickenSEM1.png" height="276px" width="600px"/> Decomposition of SNP effects <img src="chickenSEM2.png" height="176px" width="500px"/> --- # Manhattan plots of BW (overall, direct, and indirect SNP effects) <img src="chickenSEM6.png" height="426px" width="560px"/> --- # More details about Chicken SEM-GWAS <img src="frontiers.png" height="400px" width="630px"/> --- # Rice data - Drought data - Rice diversity panel - Projected shoot area - Root biomass - Water use efficiency - Water use Inferred trait network structure <img src="riceSEM1.png" height="326px" width="460px"/> --- # Inferred trait network structure <img src="riceSEM3.png" height="426px" width="700px"/> --- # Structural equation model GWAS (SEM-GWAS) `\begin{align*} \mathbf{y} =\boldsymbol{\Lambda} \mathbf{y} + \mathbf{ws} + \mathbf{Zg} + \boldsymbol{\epsilon} \end{align*}` <img src="riceSEM2.png" height="426px" width="600px"/> --- # Structural equation coefficients <img src="riceSEM4.png" height="506px" width="700px"/> --- # Projected shoot area `\begin{align*} \text{Direct}_{s_j \rightarrow y_{1_{\text{PSA}}}} &= s_{j(y_{1_{\text{PSA}}})} \\ \text{Total}_{s_j \rightarrow y_{1_{\text{PSA}}}} &= \text{Direct}_{s_j \rightarrow y_{1_{\text{PSA}}}}\\ &= s_{j(y_{1_{\text{PSA}}})} \end{align*}` <img src="riceSEM5.png" height="406px" width="700px"/> --- # Root biomass `\begin{align*} \text{Direct}_{s_j \rightarrow y_{2_{\text{RB}}}} &=s_{j(y_{2_{\text{RB}}})} \\ \text{Total}_{s_j \rightarrow y_{2_{\text{RB}}}} &= \text{Direct}_{s_j \rightarrow y_{2_{\text{RB}}}}\\ &= s_{j(y_{2_{\text{RB}}})} \end{align*}` <img src="riceSEM6.png" height="406px" width="700px"/> --- # Water use `\begin{align*} \text{Direct}_{s_j \rightarrow y_{3_{\text{WU}}}} &=s_{j(y_{3_{\text{WU}}})} \\ \text{Indirect(1)}_{s_j \rightarrow y_{3_{\text{WU}}}} &= \lambda_{13}s_{j(y_{1_{\text{PSA}}})} \\ \text{Indirect(2)}_{s_j \rightarrow y_{3_{\text{WU}}}} &= \lambda_{23}s_{j(y_{2_{\text{RB}}})} \\ \text{Total}_{s_j \rightarrow y_{2_{\text{WU}}}} &= \text{Direct}_{s_j \rightarrow y_{2_{\text{WU}}}} + \text{Indirect(1)}_{s_j \rightarrow y_{3_{\text{WU}}}} + \text{Indirect(2)}_{s_j \rightarrow y_{3_{\text{WU}}}}\\ &= s_{j(y_{3_{\text{WU}}})} + \lambda_{13}s_{j(y_{1_{\text{PSA}}})} + \lambda_{23}s_{j(y_{2_{\text{RB}}})} \end{align*}` <img src="riceSEM7.png" height="336px" width="700px"/> --- # Water use efficiency `\begin{align*} \text{Direct}_{s_j \rightarrow y_{4_{\text{WUE}}}} &=s_{j(y_{4_{\text{WUE}}})} \\ \text{Indirect(1)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{14}s_{j(y_{1_{\text{PSA}}})} \\ \text{Indirect(2)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{34}s_{j(y_{3_{\text{WU}}})} \\ \text{Indirect(3)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{13}\lambda_{34}s_{j(y_{1_{\text{PSA}}})} \\ \text{Indirect(4)}_{s_j \rightarrow y_{4_{\text{WUE}}}} &= \lambda_{23}\lambda_{34}s_{j(y_{2_{\text{RB}}})} \\ \text{Total}_{S_j \rightarrow y_{4_{\text{WUE}}}} &= \text{Direct}_{s_j \rightarrow y_{4_{\text{WUE}}}} + \text{Indirect(1)}_{s_j \rightarrow y_{4_{\text{WUE}}}} \\ &+ \text{Indirect(2)}_{s_j \rightarrow y_{4_{\text{WUE}}}} + \text{Indirect(3)}_{s_j \rightarrow y_{4_{\text{WUE}}}} \\ &+ \text{Indirect(4)}_{s_j \rightarrow y_{4_{\text{WUE}}}}\\ &= s_{j(y_{4_{\text{WUE}}})} + \lambda_{14}s_{j(y_{1_{\text{PSA}}})} + \lambda_{34}s_{j(y_{3_{\text{WU}}})} \\ &+ \lambda_{13}\lambda_{34}s_{j(y_{1_{\text{PSA}}})} + \lambda_{23}\lambda_{34}s_{j(y_{2_{\text{RB}}})} \end{align*}` --- # Water use efficiency <img src="riceSEM8.png" height="536px" width="700px"/> --- # Preprint <img src="SEMpreprint.png" height="436px" width="650px"/> --- # Ongoing SEM-GWAS projects - Italian Brown Swiss cattle (University of Padua) - Milk yield - Milk pH - Milk lactose - Somatic cell score - Non-casein N