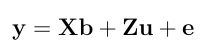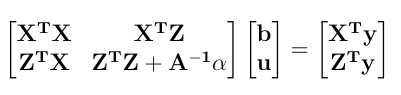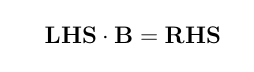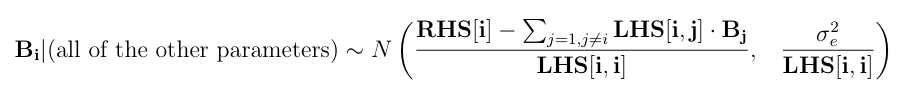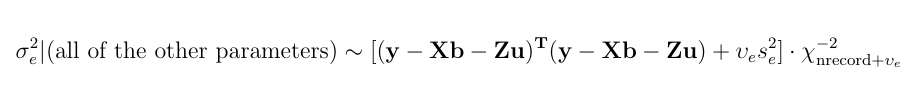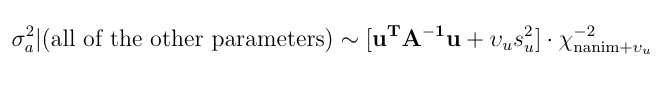Here is a numerical application from Mrode (2005), example 12.1, which deals with the pre-weaning gain (WWG) of beef calves.
> # response
> y <- c(4.5,2.9,3.9,3.5,5.0)
>
> # pedigree
> s <- c(0,0,0,1,3,1,4,3)
> d <- c(0,0,0,0,2,2,5,6)
> Ainv <- quass(s,d)
>
> # X matrix
> X <- matrix(c(1,0,0,1,1,0,1,1,0,0), nrow=5, ncol=2)
> X
[,1] [,2]
[1,] 1 0
[2,] 0 1
[3,] 0 1
[4,] 1 0
[5,] 1 0
> # Z matrix
> z1 <- matrix(0, ncol=3, nrow=5)
> z2 <- matrix(c(1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1), ncol=5)
> Z <- cbind(z1,z2)
> Z
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 0 0 0 1 0 0 0 0
[2,] 0 0 0 0 1 0 0 0
[3,] 0 0 0 0 0 1 0 0
[4,] 0 0 0 0 0 0 1 0
[5,] 0 0 0 0 0 0 0 1
Set the initials of B (b and u) to zero, the environmental variance to 40, the additive genetic variance to 20 and assume "naive" ignorance improper priors for the variance components such that ve = va = 0 and s2e = s2e = 0
> inits <- c(0,0,0,0,0,0,0,0,0,0)
> varE <- 40
> varA <- 20
> ve <- 0
> va <- 0
> s2e <- 0
> s2a <- 0
Further, allow 2000 "burnin" samples. Then collect 5000 samples after the burnin period. Define the disp option as TRUE.
> burnin <- 2000
> N <- 5000
> disp <- TRUE