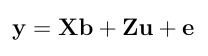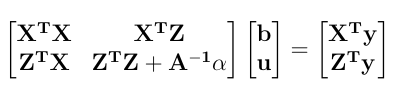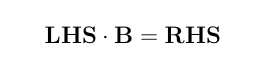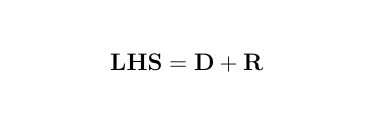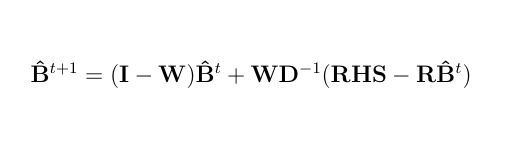Here is a numerical application from Mrode (2005), example 3.1, concerning the pre-weaning gain (WWG) of beef calves.
> # response
> y <- c(4.5,2.9,3.9,3.5,5.0)
>
> # pedigree
> s <- c(0,0,0,1,3,1,4,3)
> d <- c(0,0,0,0,2,2,5,6)
> Ainv <- quass(s,d)
>
> # variance component ratio
> vcr = 40/20
>
> # X matrix
> X <- matrix(c(1,0,0,1,1,0,1,1,0,0), nrow=5, ncol=2)
> X
[,1] [,2]
[1,] 1 0
[2,] 0 1
[3,] 0 1
[4,] 1 0
[5,] 1 0
> # Z matrix
> z1 <- matrix(0, ncol=3, nrow=5)
> z2 <- matrix(c(1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1), ncol=5)
> Z <- cbind(z1,z2)
> Z
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 0 0 0 1 0 0 0 0
[2,] 0 0 0 0 1 0 0 0
[3,] 0 0 0 0 0 1 0 0
[4,] 0 0 0 0 0 0 1 0
[5,] 0 0 0 0 0 0 0 1
> Xpy <- t(X)%*%y
> Zpy <- t(Z)%*%y
> XpX <- t(X)%*% X
> XpZ <- t(X)%*%Z
> ZpX <- t(Z)%*%X
> ZpZ <- t(Z)%*%Z
>
> LHS <- rbind( cbind(XpX, XpZ), cbind(ZpX, ZpZ+Ainv*vcr))
> RHS <- c(Xpy, Zpy)
The relaxation factor has been chosen as 1.0 for the fixed effects and 0.8 for the random effects.
Initial values included the mean yield for fixed effects. For animals with phenotypic records, initial values were the deviation of yields from the mean yield of their respective fixed effects and zero for animals without phenotypic records.
> inits <- c(4.333,3.400,0,0,0,0.167,-0.5,0.5,-0.833,0.667)
> w <- c(1,1,rep(0.8,8))
> disp = FALSE