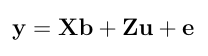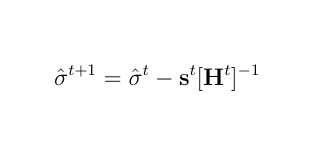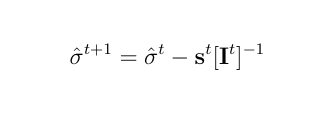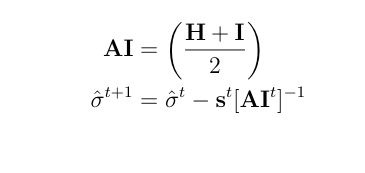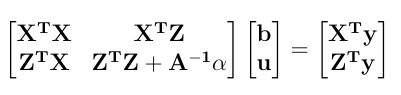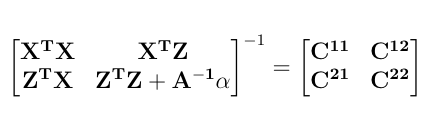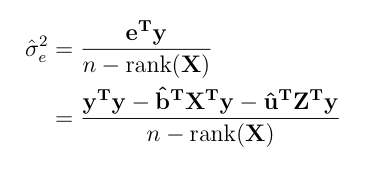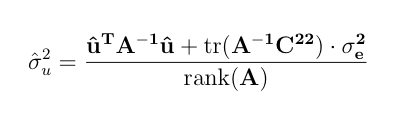- Given the components of the MME, estimate variance components by REML using Newton Raphson framework. Consider the following univariate mixed linear model.
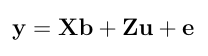 The Newton Raphson method works as the next equation and repeat this process untill it achives convergence.
The Newton Raphson method works as the next equation and repeat this process untill it achives convergence.
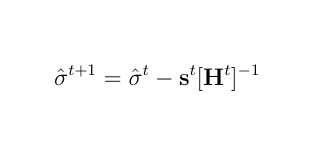 where s is the score vector and H is the Hessian matrix for the log-likelihood of variance components.
where s is the score vector and H is the Hessian matrix for the log-likelihood of variance components.
-
Here is a numerical application where the data is from Mrode (2005), example 11.6, concerning the pre-weaning gain (WWG) of beef calves.
> # response
> y <- c(2.6,0.1,1.0,3.0,1.0)
>
> # pedigree
> s <- c(0,0,0,1,3,1,4,3)
> d <- c(0,0,0,0,2,2,5,6)
> A <- createA(s,d)
>
> # X matrix
> X <- matrix(c(1,0,0,1,1,0,1,1,0,0), nrow=5, ncol=2)
> X
[,1] [,2]
[1,] 1 0
[2,] 0 1
[3,] 0 1
[4,] 1 0
[5,] 1 0
>
> # Z matrix
> z1 <- matrix(0, ncol=3, nrow=5)
> z2 <- matrix(c(1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1), ncol=5)
> Z <- cbind(z1,z2)
> Z
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 0 0 0 1 0 0 0 0
[2,] 0 0 0 0 1 0 0 0
[3,] 0 0 0 0 0 1 0 0
[4,] 0 0 0 0 0 0 1 0
[5,] 0 0 0 0 0 0 0 1
In this particular design, the Hessian Matrix is going to be singular and unable to take inverse. Thus, we will add one data point.
> y <- c(y, 2.0)
> addX <- matrix(c(0,1), ncol=2)
> X <- rbind(X,addX)
> X
[,1] [,2]
[1,] 1 0
[2,] 0 1
[3,] 0 1
[4,] 1 0
[5,] 1 0
[6,] 0 1
> addZ <- matrix(c(0,0,0,0,0,1,0,0), ncol=8)
> Z <- rbind(Z, addZ)
> Z
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 0 0 0 1 0 0 0 0
[2,] 0 0 0 0 1 0 0 0
[3,] 0 0 0 0 0 1 0 0
[4,] 0 0 0 0 0 0 1 0
[5,] 0 0 0 0 0 0 0 1
[6,] 0 0 0 0 0 1 0 0
Our initials for the variance components will be:
> initE <- 0.4
> initU <- 0.2
> disp <- TRUE
After 18 iterations, Newton Raphson gives following estimates.
> nrReml(A, y, X, Z, initE, initU, disp)
iteration 1
sig2U 0.3055028
sig2E 0.4688081
iteration 2
sig2U 0.4567926
sig2E 0.4962741
iteration 3
sig2U 0.6235681
sig2E 0.4954533
.
.
snip
.
.
iteration 16
sig2U 1.128275
sig2E 0.4891807
iteration 17
sig2U 1.128592
sig2E 0.489179
iteration 18
sig2U 1.128775
sig2E 0.489178
$sigma2U
[1] 1.128775
$sigma2E
[1] 0.489178
- Given the components of MME, estimate variance components by the EM Algorithm. Consider the following univariate mixed linear model.
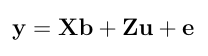 In the Mixed Model Equation form:
In the Mixed Model Equation form:
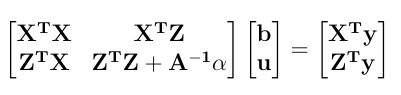 Further, let the inverse of the left hand side be:
Further, let the inverse of the left hand side be:
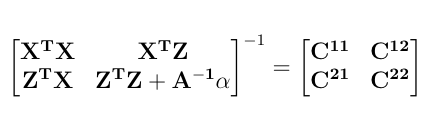 The EM Algorithm can be implemented by following successive iterations.
The EM Algorithm can be implemented by following successive iterations.
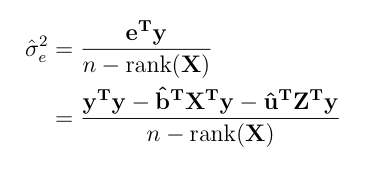
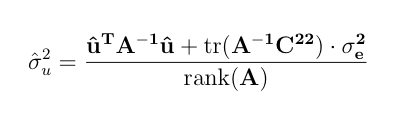
-
An example is taken from Mrode (2005), 11.6, concerning the pre-weaning gain (WWG) of beef calves.
> # response
> y <- c(2.6,0.1,1.0,3.0,1.0)
>
> # pedigree
> s <- c(0,0,0,1,3,1,4,3)
> d <- c(0,0,0,0,2,2,5,6)
> Ainv <- quass(s,d)
>
> # X matrix
> X <- matrix(c(1,0,0,1,1,0,1,1,0,0), nrow=5, ncol=2)
> X
[,1] [,2]
[1,] 1 0
[2,] 0 1
[3,] 0 1
[4,] 1 0
[5,] 1 0
>
> # Z matrix
> z1 <- matrix(0, ncol=3, nrow=5)
> z2 <- matrix(c(1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1,0,0,0,0,0,1), ncol=5)
> Z <- cbind(z1,z2)
> Z
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 0 0 0 1 0 0 0 0
[2,] 0 0 0 0 1 0 0 0
[3,] 0 0 0 0 0 1 0 0
[4,] 0 0 0 0 0 0 1 0
[5,] 0 0 0 0 0 0 0 1
As in Mrode (2005), set the initial values for the residual variance to 0.4 and set the additive variance to 0.2.
> # initial variance components
> initE <- 0.4
> initU <- 0.2
>
> disp = TRUE
The EM algorithm gives the following estimates after 906 iterations.
> emReml(Ainv, y, X, Z, initE, initU, disp)
iteration 1
sig2E 0.6425753
sig2U 0.2124631
iteration 2
sig2E 0.7061095
sig2U 0.2145396
iteration 3
sig2E 0.7174604
sig2U 0.2153153
.
.
snip
.
.
iteration 904
sig2E 0.4848573
sig2U 0.5493628
iteration 905
sig2E 0.4848473
sig2U 0.5493777
iteration 906
sig2E 0.4848373
sig2U 0.5493925
$sigma2E
[,1]
[1,] 0.4848373
$sigma2U
[,1]
[1,] 0.5493925